Below are highlights from Technology Research and Development 2 projects covering PET/MR reconstruction and estimations.
ACT: Semi-supervised domain-adaptive medical image segmentation with asymmetric co-training
The article discusses a new method for improving the accuracy of medical image segmentation, specifically in cases where there is a significant difference between the labeled source domain and the unlabeled target domain. The proposed method is called semi-supervised domain adaptation (SSDA), which involves the use of both labeled source and target domain data, as well as unlabeled target data in a unified manner. The authors present a novel asymmetric co-training (ACT) framework to integrate these subsets, which yielded marked improvements over existing methods in cross-modality brain tumor MRI segmentation tasks.
Abstract

(e.g., T2-weighted to T1-weighted MRI) image segmentation. N
Unsupervised domain adaptation (UDA) has been vastly explored to alleviate domain shifts between source and target domains, by applying a well-performed model in an unlabeled target domain via supervision of a labeled source domain. Recent literature, however, has indicated that the performance is still far from satisfactory in the presence of significant domain shifts. Nonetheless, delineating a few target samples is usually manageable and particularly worthwhile, due to the substantial performance gain. Inspired by this, we aim to develop semi-supervised domain adaptation (SSDA) for medical image segmentation, which is largely underexplored. We, thus, propose to exploit both labeled source and target domain data, in addition to unlabeled target data in a unified manner. Specifically, we present a novel asymmetric co-training (ACT) framework to integrate these subsets and avoid the domination of the source domain data. Following a divide-and-conquer strategy, we explicitly decouple the label supervisions in SSDA into two asymmetric sub-tasks, including semi-supervised learning (SSL) and UDA, and leverage different knowledge from two segmentors to take into account the distinction between the source and target label supervisions. The knowledge learned in the two modules is then adaptively integrated with ACT, by iteratively teaching each other, based on the confidence-aware pseudo-label. In addition, pseudo label noise is well-controlled with an exponential MixUp decay scheme for smooth propagation. Experiments on cross-modality brain tumor MRI segmentation tasks using the BraTS18 database showed, even with limited labeled target samples, ACT yielded marked improvements over UDA and state-of-the-art SSDA methods and approached an “upper bound” of supervised joint training.
X. Liu, et. al. pp 66-76 MICCAI 2022.
Unsupervised Black-Box Model Domain Adaptation for Brain Tumor Segmentation
This article discusses unsupervised domain adaptation (UDA) that transfers domain knowledge from a labeled source domain to unlabeled target domains. The proposed framework suggests a black-box segmentation model trained in the source domain only, using a knowledge distillation scheme to learn target-specific representations, and regularizing the confidence of the labels in the target domain via unsupervised entropy minimization. The framework was validated on multiple datasets and deep learning backbones, demonstrating potential for use in challenging clinical settings.
Abstract
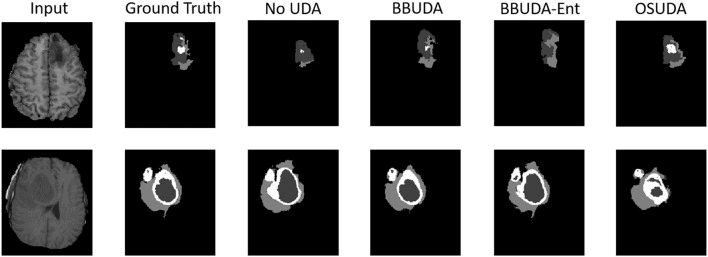
Unsupervised domain adaptation (UDA) is an emerging technique that enables the transfer of domain knowledge learned from a labeled source domain to unlabeled target domains, providing a way of coping with the difficulty of labeling in new domains. The majority of prior work has relied on both source and target domain data for adaptation. However, because of privacy concerns about potential leaks in sensitive information contained in patient data, it is often challenging to share the data and labels in the source domain and trained model parameters in cross-center collaborations. To address this issue, we propose a practical framework for UDA with a black-box segmentation model trained in the source domain only, without relying on source data or a white-box source model in which the network parameters are accessible. In particular, we propose a knowledge distillation scheme to gradually learn target-specific representations. Additionally, we regularize the confidence of the labels in the target domain via unsupervised entropy minimization, leading to performance gain over UDA without entropy minimization. We extensively validated our framework on a few datasets and deep learning backbones, demonstrating the potential for our framework to be applied in challenging yet realistic clinical settings.
X. Liu et. al., Front. Neurosci., 02 June 2022,Volume 16
Posterior estimation using deep learning: a simulation study of compartmental modeling in dynamic positron emission tomography
This study aims to use deep learning to estimate uncertainties in medical images, which are often treated as deterministic. The researchers developed two deep neural networks based on conditional variational auto-encoder (CVAE) to implement a variational Bayesian inference framework, which estimates posterior distributions of imaging parameters. Their simulation study of dynamic brain PET imaging showed that their proposed CVAE-dual-encoder and CVAE-dual-decoder approaches yield good results and can be adapted for other medical imaging applications.
Abstract
Background and Purpose
In medical imaging, images are usually treated as deterministic, while their uncertainties are largely underexplored. This work aims at using deep learning to efficiently estimate posterior distributions of imaging parameters, which in turn can be used to derive the most probable parameters as well as their uncertainties.
Methods
Our deep learning-based approaches are based on a variational Bayesian inference framework, which is implemented using two different deep neural networks based on conditional variational auto-encoder (CVAE), CVAE-dual-encoder, and CVAE-dual-decoder. The conventional CVAE framework, that is, CVAE-vanilla, can be regarded as a simplified case of these two neural networks. We applied these approaches to a simulation study of dynamic brain PET imaging using a reference region-based kinetic model.
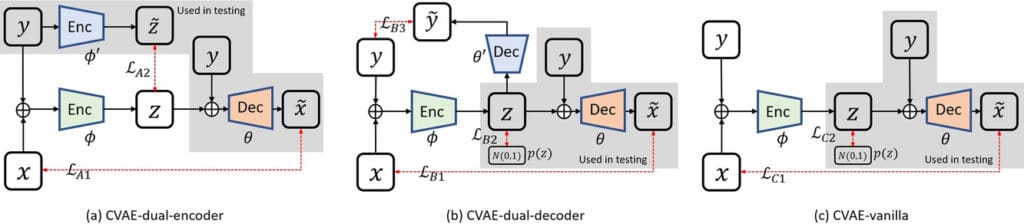
Results
In the simulation study, we estimated posterior distributions of PET kinetic parameters given a measurement of the time–activity curve. Our proposed CVAE-dual-encoder and CVAE-dual-decoder yield results that are in good agreement with the asymptotically unbiased posterior distributions sampled by Markov Chain Monte Carlo (MCMC). The CVAE-vanilla can also be used for estimating posterior distributions, although it has an inferior performance to both CVAE-dual-encoder and CVAE-dual-decoder.
Conclusions
We have evaluated the performance of our deep learning approaches for estimating posterior distributions in dynamic brain PET. Our deep learning approaches yield posterior distributions, which are in good agreement with unbiased distributions estimated by MCMC. All these neural networks have different characteristics and can be chosen by the user for specific applications. The proposed methods are general and can be adapted to other problems.
Liu X, et. al., Med Phys. 2023 Mar;50(3):1539-1548.
Free-breathing 3D cardiac T mapping with transmit B correction at 3T
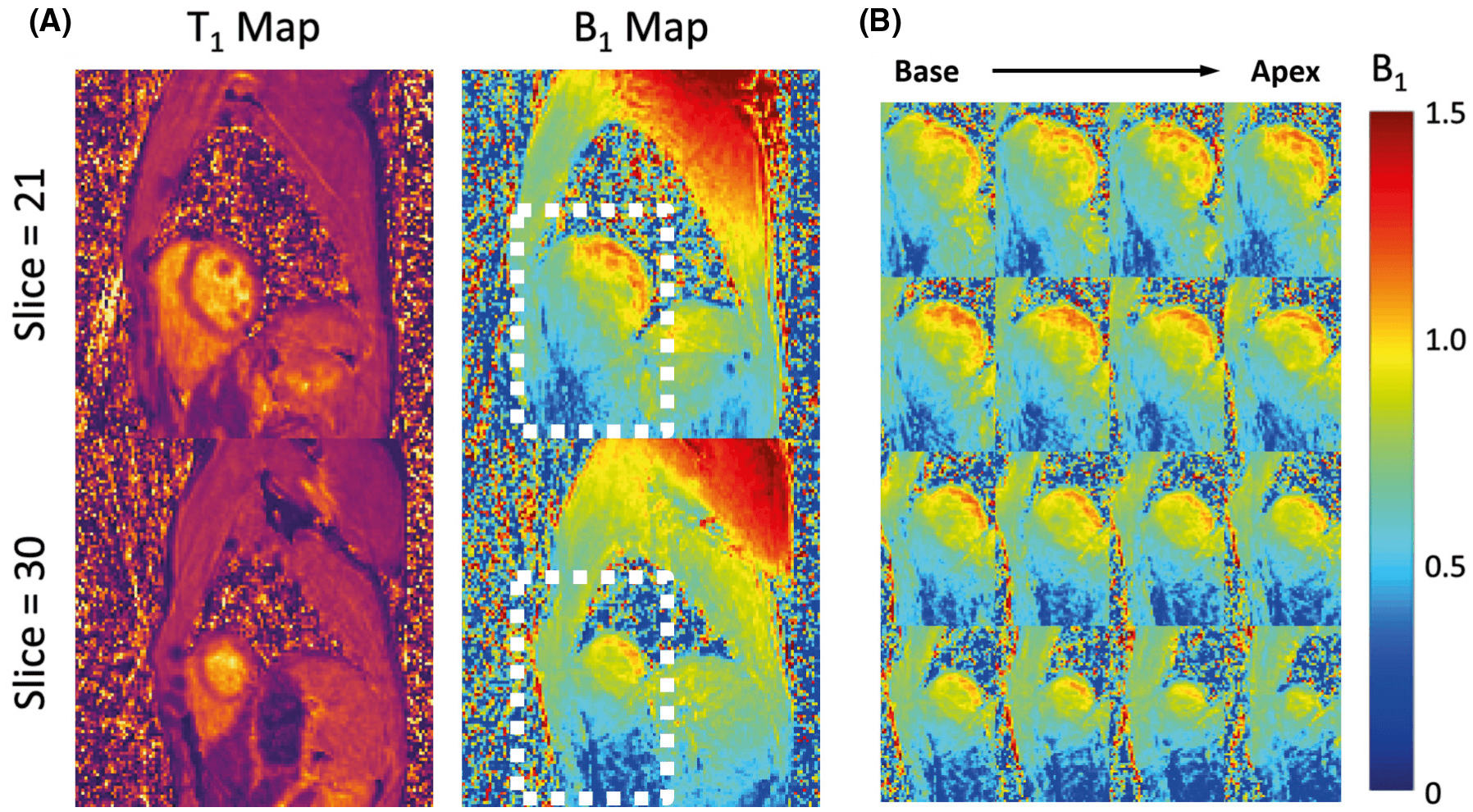
Purpose: To develop a cardiac T1 mapping method for free-breathing 3D T1 mapping of the whole heart at 3 T with transmit B1 ( B+1 ) correction.
Methods: A free-breathing, electrocardiogram-gated inversion-recovery sequence with spoiled gradient-echo readout was developed and optimized for cardiac T1 mapping at 3 T. High-frame-rate dynamic images were reconstructed from sparse (k,t)-space data acquired along a stack-of-stars trajectory using a subspace-based method for accelerated imaging. Joint T1 and flip-angle estimation was performed in T1 mapping to improve its robustness to B1+ inhomogeneity. Subject-specific timing of data acquisition was used in the estimation to account for natural heart-rate variations during the imaging experiment.
Results: Simulations showed that accuracy and precision of T1 mapping can be improved with joint T1 and flip-angle estimation and optimized electrocardiogram-gated spoiled gradient echo-based inversion-recovery acquisition scheme. The phantom study showed good agreement between the T1 maps from the proposed method and the reference method. Three-dimensional cardiac T1 maps (40 slices) were obtained at a 1.9-mm in-plane and 4.5-mm through-plane spatial resolution from healthy subjects (n = 6) with an average imaging time of 14.2 ± 1.6 minutes (heartbeat rate: 64.2 ± 7.1 bpm), showing myocardial T1 values comparable to those obtained from modified Look-Locker inversion recovery. The proposed method generated B+1 maps with spatially smooth variation showing 21%-32% and 11%-15% variations across the septal-lateral and inferior-anterior regions of the myocardium in the left ventricle.
Conclusion: The proposed method allows free-breathing 3D T1 mapping of the whole heart with transmit B1 correction in a practical imaging time.
Han PK, et. al., Magn Reson Med 2022 87 (4):1832-1845
Arterial spin labeling MR image denoising and reconstruction
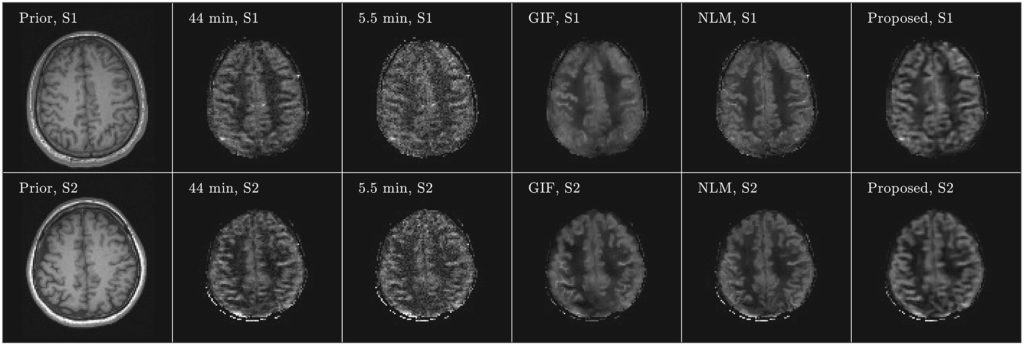
Arterial spin labeling (ASL) imaging is a powerful magnetic resonance imaging technique that allows to quantitatively measure blood perfusion non‐invasively, which has great potential for assessing tissue viability in various clinical settings. For this work we created an unsupervised deep learning‐based image denoising and reconstruction framework to improve the SNR and accelerate the imaging speed of high resolution ASL imaging. The proposed framework does not require any prior training pairs but only the subject’s own anatomical prior, such as T1‐weighted images, as network input.
Both qualitative and quantitative analyses demonstrate the superior performance of the proposed framework over the reference methods in a study with three healthy volunteers.
Gong K, Han P, El Fakhri G, Ma C, Li Q. NMR Biomed 2022 35 (4):e4224
PET imaging of neurotransmission using direct parametric reconstruction
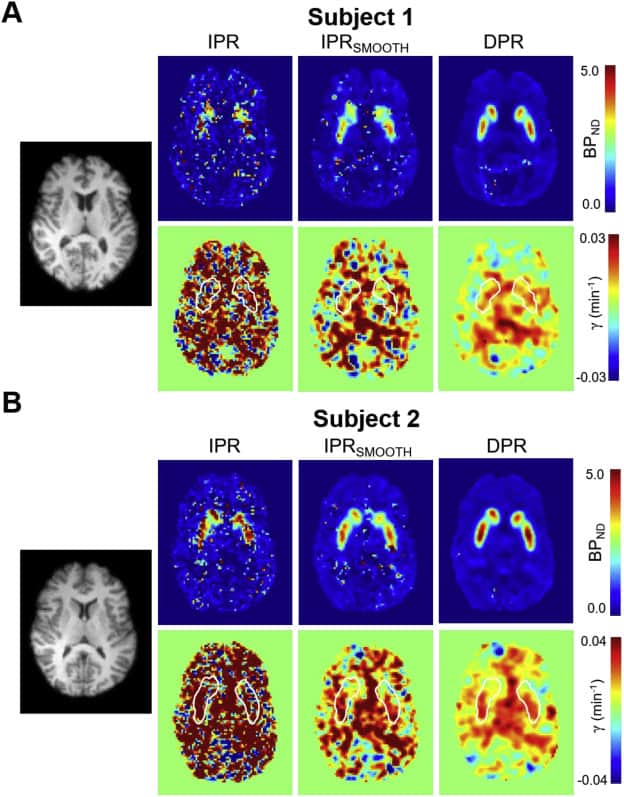
Receptor ligand-based dynamic Positron Emission Tomography (PET) permits the measurement of neurotransmitter release in the human brain. For single-scan paradigms, the conventional method of estimating changes in neurotransmitter levels relies on fitting a pharmacokinetic model to activity concentration histories extracted after PET image reconstruction (indirect parametric reconstruction, IPR). However, due to the statistical fluctuations of activity concentration data at the voxel scale, parametric images computed using this approach often exhibit low signal-to-noise ratio, impeding characterization of neurotransmitter release. Numerous studies have shown that direct parametric reconstruction (DPR) approaches, which combine image reconstruction and kinetic analysis in a unified framework, can improve the signal-to-noise ratio of parametric mapping. However, there is little experience with DPR in imaging of neurotransmission and the performance of the approach in this application has not been evaluated before in humans. In this report, we present and evaluate a DPR methodology that computes 3-D distributions of ligand transport, binding potential (BPND) and neurotransmitter release magnitude (γ) from a dynamic sequence of PET sinograms. The technique employs the linear simplified reference region model (LSRRM) of Alpert et al. (2003), which represents an extension of the simplified reference region model that incorporates time-varying binding parameters due to radioligand displacement by release of neurotransmitter. Estimation of parametric images is performed by gradient-based optimization of a Poisson log-likelihood function incorporating LSRRM kinetics and accounting for the effects of head movement, attenuation, detector sensitivity, random and scattered coincidences. A 11C-raclopride simulation study showed that the proposed approach substantially reduces the bias and variance of voxel-wise γ estimates as compared to standard methods. Moreover, simulations showed that detection of release could be made more reliable and/or conducted using a smaller sample size using the proposed DPR estimator. Likewise, images of BPND computed using DPR had substantially improved bias and variance properties. Application of the method in human subjects was demonstrated using 11C-raclopride dynamic scans and a reward task, confirming the improved quality of the estimated parametric images using the proposed approach.
Y. Petibon, et. al. NeuroImage, Volume 221, 1 November 2020, 117154
Penalized Parametric PET Image Estimation Using Local Linear Fitting
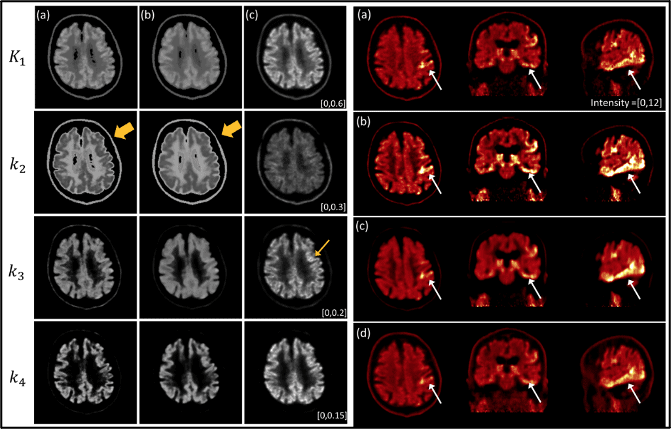
Positron emission tomography (PET) is a powerful imaging tool for quantifying physiological information. Particularly, parametric PET imaging has been increasingly investigated because the voxel-wise spatial distribution of tracer kinetics can be achieved, which is suitable for identifying heterogeneous tracer uptake. Our goal is to provide a fast and accurate parametric imaging method by utilizing the full-dose image. In this article, we propose a novel penalized parametric imaging in which dynamic images are reconstructed frame by frame, and then voxel-wise kinetic parameters are iteratively estimated by a kinetic-domain penalty using a local linear fitting (LLF) that utilizes a full-dose static image as a prior in the penalty. In our optimization, a dual-domain approximation is derived to efficiently split suboptimizations for image and kinetic parameter separately, which can simplify suboptimizations including the nonlinear fitting of the two-tissue compartment model (2-TCM). In addition, we further investigate the feasibility of image-driven input function extraction by comparing with the standard blood sampling. We evaluated the performance of the proposed method using simulation and patient study with [ 18 F]MK6240 tau scan data of two subjects, and results are compared with the conventional method and the Logan graphical model. We demonstrate that the proposed method outperforms the conventional parametric imaging methods.
K. Kim, et. al. IEEE Transactions on Radiation and Plasma Medical Sciences, doi: 10.1109/TRPMS.2020; 4(6):750-758
Preclinical Validation of a Single-Scan Rest/Stress Imaging Technique for 13N-Ammonia Positron Emission Tomography Cardiac Perfusion Studies.
Background: We previously proposed a technique for quantitative measurement of rest and stress absolute myocardial blood flow (MBF) using a 2-injection single-scan imaging session. Recently, we validated the method in a pig model for the long-lived radiotracer 18F-Flurpiridaz with adenosine as a pharmacological stressor. The aim of the present work is to validate our technique for 13NH3.
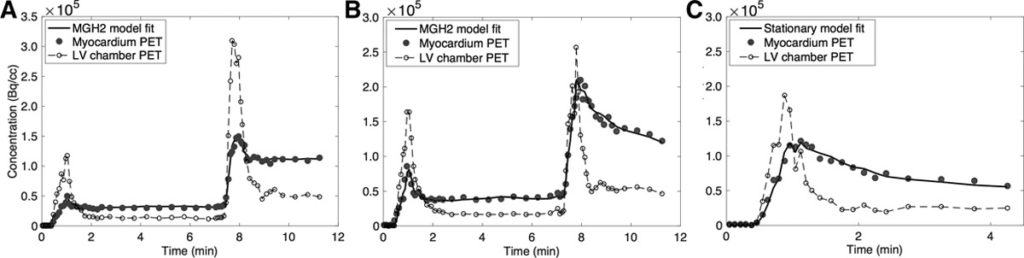
Methods: Nine studies were performed in 6 pigs; 5 studies were done in the native state and 4 after infarction of the left anterior descending artery. Each study consisted of 3 dynamic scans: a 2-injection rest-rest single-scan acquisition (scan A), a 2-injection rest/stress single-scan acquisition (scan B), and a conventional 1-injection stress acquisition (scan C). Variable doses of adenosine combined with dobutamine were administered to induce a wide range of MBF. The 2-injection single-scan measurements were fitted with our nonstationary kinetic model (MGH2). In 4 studies, 13NH3 injections were paired with microsphere injections. MBF estimates obtained with our method were compared with those obtained with the standard method and with microspheres. We used a model-based method to generate separate rest and stress perfusion images.
Results: In the absence of stress (scan A), the MBF values estimated by MGH2 were nearly the same for the 2-radiotracer injections (mean difference: 0.067±0.070 mL·min-1·cc-1, limits of agreement: [-0.070 to 0.204] mL·min-1·cc-1), showing good repeatability. Bland-Altman analyses demonstrated very good agreement with the conventional method for both rest (mean difference: -0.034±0.035 mL·min-1·cc-1, limits of agreement: [-0.103 to 0.035] mL·min-1·cc-1) and stress (mean difference: 0.057±0.361 mL·min-1·cc-1, limits of agreement: [-0.651 to 0.765] mL·min-1·cc-1) MBF measurements. Positron emission tomography and microsphere MBF measurements correlated closely. Very good quality perfusion images were obtained.
Conclusions: This study provides in vivo validation of our single-scan rest-stress method for 13NH3 measurements. The 13NH3 rest/stress myocardial perfusion imaging procedure can be compressed into a single positron emission tomography scan session lasting less than 15 minutes.
NJ Guehl, et. al. Circ Cardiovasc Imaging. 2020 Jan;13(1):e009407. doi: 10.1161/CIRCIMAGING.119.009407.
PET image deblurring and super-resolution with an MR-based joint entropy prior
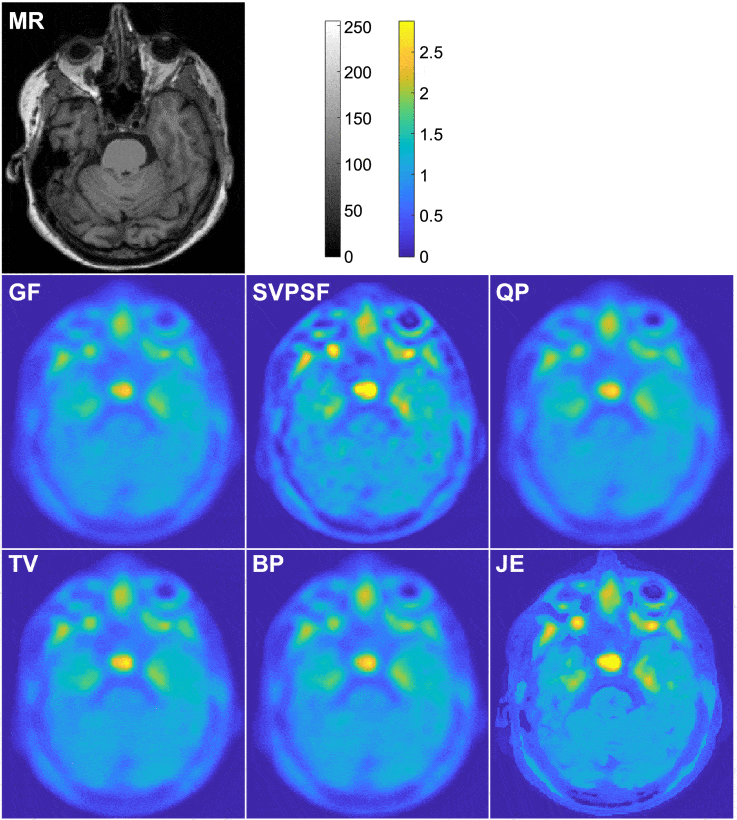
The intrinsically limited spatial resolution of PET confounds image quantitation. This paper presents an image deblurring and super-resolution framework for PET using anatomical guidance provided by high-resolution MR images. The framework relies on image-domain post-processing of already-reconstructed PET images by means of spatially-variant deconvolution stabilized by an MR-based joint entropy penalty function. The method is validated through simulation studies based on the BrainWeb digital phantom, experimental studies based on the Hoffman phantom, and clinical neuroimaging studies pertaining to aging and Alzheimer’s disease. The developed technique was compared with direct deconvolution and deconvolution stabilized by a quadratic difference penalty, a total variation penalty, and a Bowsher penalty.
The BrainWeb simulation study showed improved image quality and quantitative accuracy measured by contrast-to-noise ratio, structural similarity index, root-mean-square error, and peak signal-to-noise ratio generated by this technique. The Hoffman phantom study indicated noticeable improvement in the structural similarity index (relative to the MR image) and gray-to-white contrast-to-noise ratio. Finally, clinical amyloid and tau imaging studies for Alzheimer’s disease showed lowering of the coefficient of variation in several key brain regions associated with two target pathologies.
T.A. Song, et al. IEEE Trans Comput Imaging. 2019 Dec;5(4):530-539. doi: 10.1109/TCI.2019.2913287.
Gross tumor volume segmentation for head and neck cancer radiotherapy using deep dense multi-modality network
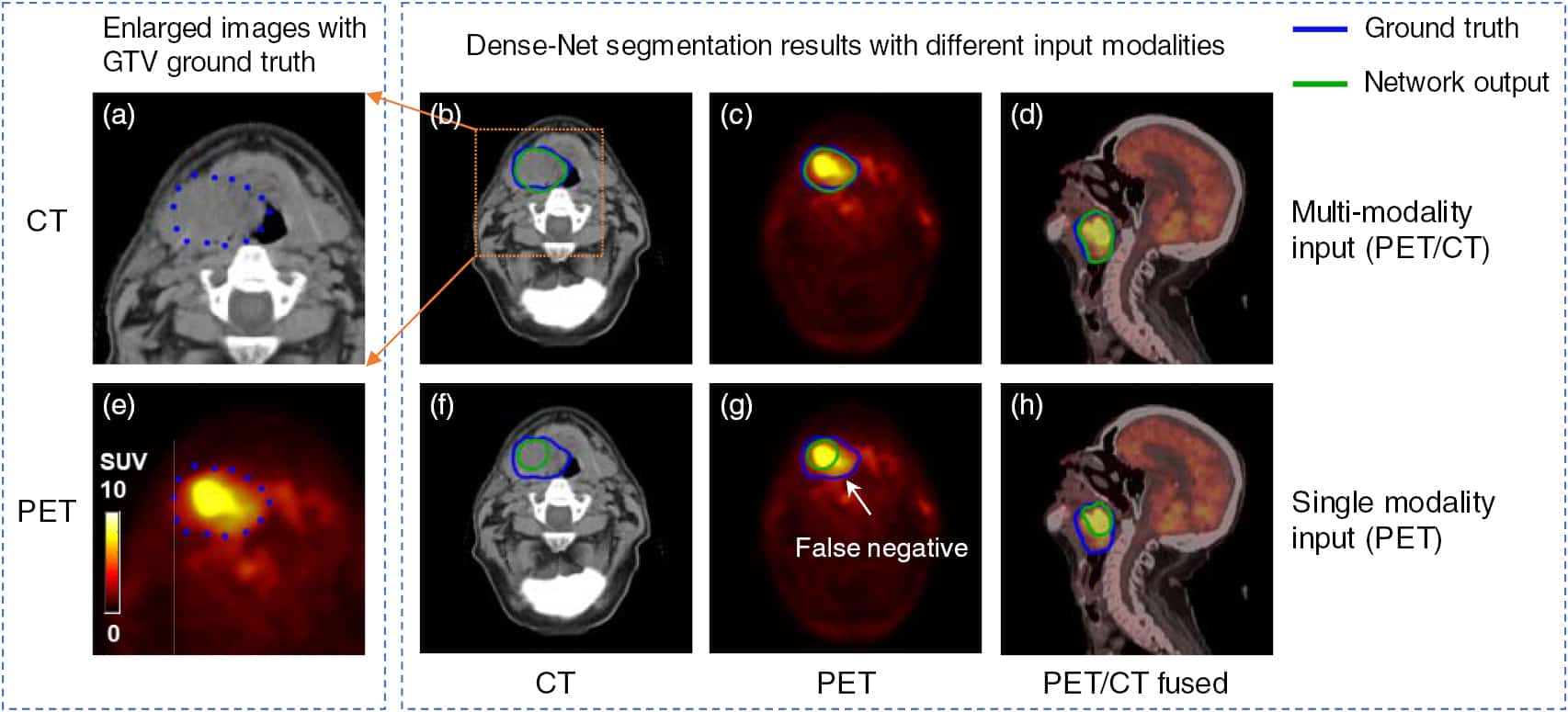
In radiation therapy, the accurate delineation of gross tumor volume (GTV) is crucial for treatment planning. However, it is challenging for head and neck cancer (HNC) due to the morphology complexity of various organs in the head, low targets to background contrast and potential artifacts on conventional planning CT images. Thus, manual delineation of GTV on anatomical images is extremely time consuming and suffers from inter-observer variability that leads to planning uncertainty. With the wide use of PET/CT imaging in oncology, complementary functional and anatomical information can be utilized for tumor contouring and bring a significant advantage for radiation therapy planning.
In this study, by taking advantage of multi-modality PET and CT images, we propose an automatic GTV segmentation framework based on deep learning for HNC. The backbone of this segmentation framework is based on 3D convolution with dense connections which enables a better information propagation and takes full advantage of the features extracted from multi-modality input images. We evaluate our proposed framework on a dataset including 250 HNC patients. Each patient receives both planning CT and PET/CT imaging before radiation therapy (RT). Manually delineated GTV contours by radiation oncologists are used as ground truth in this study. To further investigate the advantage of our proposed Dense-Net framework, we also compared with the framework using 3D U-Net which is the state-of-the-art in segmentation tasks. Meanwhile, for each frame, the performance comparison between single modality input (PET or CT image) and multi-modality input (both PET/CT) is conducted. Dice coefficient, mean surface distance (MSD), 95th-percentile Hausdorff distance (HD95) and displacement of mass centroid (DMC) are calculated for quantitative evaluation. The dataset is split into train (140 patients), validation (35 patients) and test (75 patients) groups to optimize the network. Based on the results on independent test group, our proposed multi-modality Dense-Net (Dice 0.73) shows better performance than the compared network (Dice 0.71). Furthermore, the proposed Dense-Net structure has less trainable parameters than the 3D U-Net, which reduces the prediction variability.
In conclusion, our proposed multi-modality Dense-Net can enable satisfied GTV segmentation for HNC using multi-modality images and yield superior performance than conventional methods. Our proposed method provides an automatic, fast and consistent solution for GTV segmentation and shows potentials to be generally applied for radiation therapy planning of a variety of cancer (e.g. lung, sarcoma, liver and so on).
Z. Guo, et. al. Phys Med Biol. 2019 Oct 16;64(20):205015. doi: 10.1088/1361-6560/ab440d
PET Image Reconstruction Using Deep Image Prior
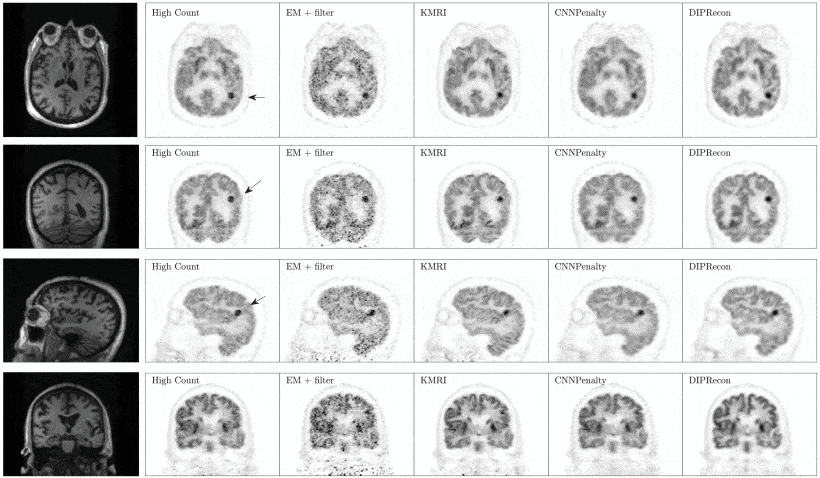
Recently, deep neural networks have been widely and successfully applied in computer vision tasks and have attracted growing interest in medical imaging. One barrier for the application of deep neural networks to medical imaging is the need for large amounts of prior training pairs, which is not always feasible in clinical practice. This is especially true for medical image reconstruction problems, where raw data are needed. Inspired by the deep image prior framework, in this paper, we proposed a personalized network training method where no prior training pairs are needed, but only the patient’s own prior information. The network is updated during the iterative reconstruction process using the patient-specific prior information and measured data. We formulated the maximum-likelihood estimation as a constrained optimization problem and solved it using the alternating direction method of multipliers algorithm. Magnetic resonance imaging guided positron emission tomography reconstruction was employed as an example to demonstrate the effectiveness of the proposed framework. Quantification results based on simulation and real data show that the proposed reconstruction framework can outperform Gaussian post-smoothing and anatomically guided reconstructions using the kernel method or the neural-network penalty.
K. Gong, et. al. IEEE Trans Med Imaging. 2019 Jul;38(7):1655-1665. doi: 10.1109/TMI.2018.2888491.
Sequence Alterations of Cortical Genes Linked to Individual Connectivity of the Human Brain.
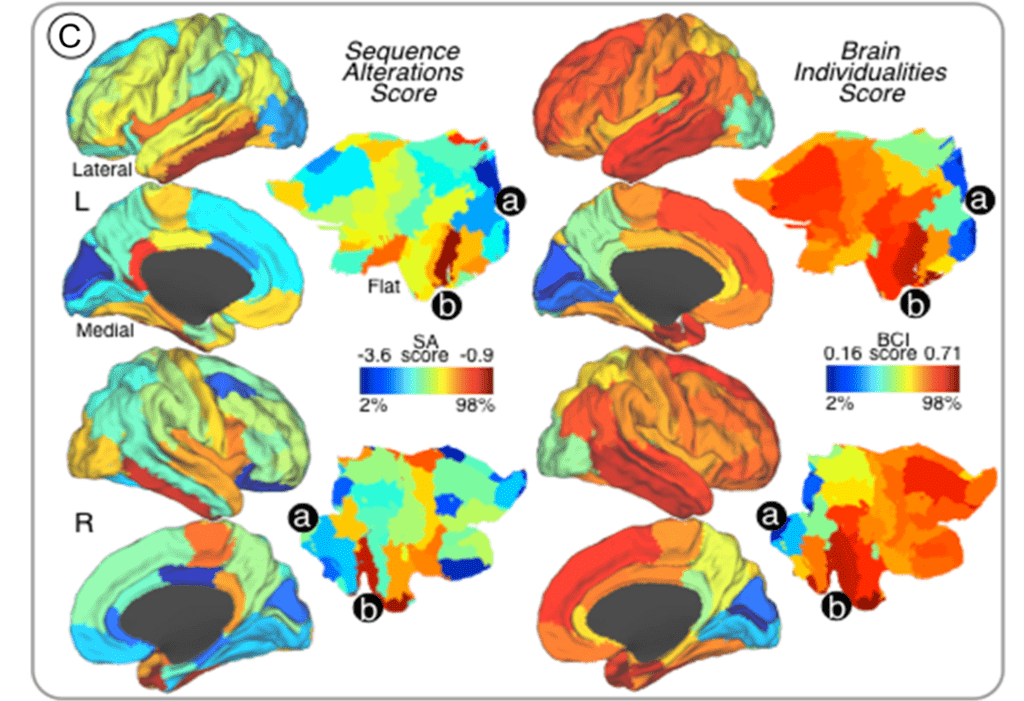
Individual differences in humans are driven by unique brain structural and functional profiles, presumably mediated in part through differential cortical gene expression. However, the relationships between cortical gene expression profiles and individual differences in large-scale neural network organization remain poorly understood.
In this study, we aimed to investigate whether the magnitude of sequence alterations in regional cortical genes mapped onto brain areas with high degree of functional connectivity variability across individuals. First, human genetic expression data from the Allen Brain Atlas was used to identify protein-coding genes associated with cortical areas, which delineated the regional genetic signature of specific cortical areas based on sequence alteration profiles. Thereafter, we identified brain regions that manifested high degrees of individual variability by using test-retest functional connectivity magnetic resonance imaging and graph-theory analyses in healthy subjects. We found that rates of genetic sequence alterations shared a distinct spatial topography with cortical regions exhibiting individualized (highly-variable) connectivity profiles. Interestingly, gene expression profiles of brain regions with highly individualized connectivity patterns and elevated number of sequence alterations are devoted to neuropeptide-signaling-pathways and chemical-synaptic-transmission.
Our findings support that genetic sequence alterations may underlie important aspects of brain connectome individualities in humans.
Q Xin, et. al. Cereb Cortex. 2019 Aug 14;29(9):3828-3835. doi: 10.1093/cercor/bhy262.
Multi-Modal Signatures of Tau Pathology, Neuronal Fiber Integrity, and Functional Connectivity in Traumatic Brain Injury
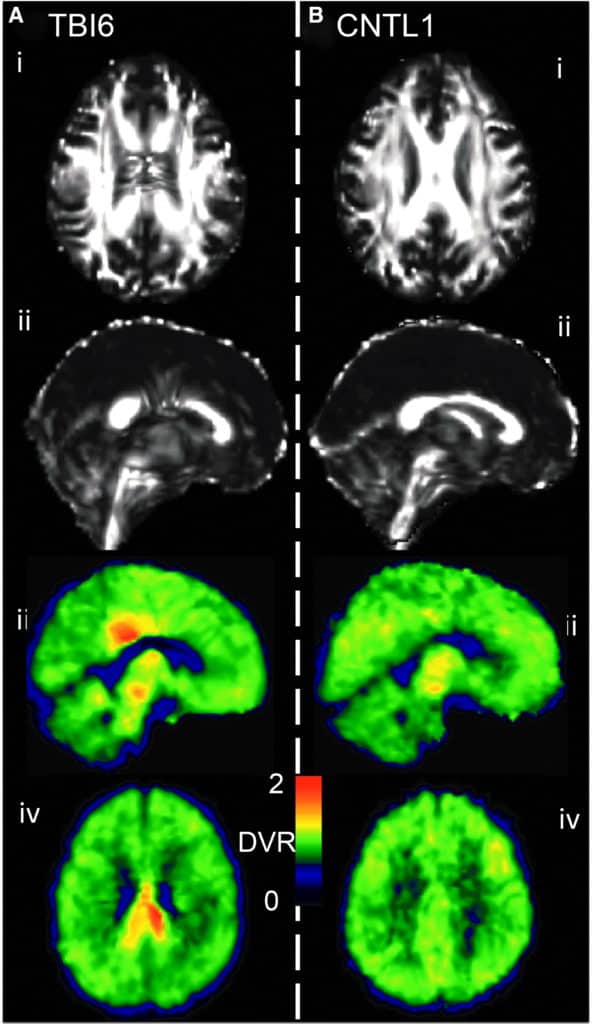
[18F]AV-1451 (aka 18F-Flortaucipir, [18F]T807) was developed for positron-emission tomography (PET) imaging of paired helical filaments of hyperphosphorylated tau, which are of interest in a range of neuropathologies, including traumatic brain injury (TBI). Magnetic resonance imaging (MRI) techniques like diffusion tensor imaging (DTI) and resting state functional connectivity assess structural and functional characteristics of the brain, complementing the molecular information that can be obtained by PET.
The goal herein was to explore the utility of such multi-modal imaging in a case series based on a population of TBI subjects. This study probes the interrelationship between tau deposition, white matter integrity, and gray matter functional connectivity across the spectrum of TBI. Nineteen subjects (11 controls, five former contact sports athletes, one automotive accident, and two with military-related injury) underwent [18F]AV-1451 PET and magnetic resonance scanning procedures. [18F]AV-1451 distribution volume ratio (DVR) was estimated using the Logan method and the cerebellum as a reference region. Diffusion tractography images and fractional anisotropy (FA) images were generated using diffusion toolkit and FSL. Resting-state functional MRI (fMRI) analysis was based on a graph theory metric, namely weighted degree centrality. TBI subjects showed greater heterogeneity in [18F]AV-1451 DVR when compared with control subjects. In a subset of TBI subjects, areas with high [18F]AV-1451 binding corresponded with increased FA and diminished white matter tract density in DTI. Functional MRI results exhibited an increase in functional connectivity, particularly among local connections, in the areas where tau aggregates were more prevalent.
In a case series of a diverse group of TBI subjects, brain regions with elevated tau burden exhibited increased functional connectivity as well as decreased white matter integrity. These findings portray molecular, microstructural, and functional corollaries of TBI that spatially coincide and can be measured in the living human brain using noninvasive neuroimaging techniques.
DW Wooten, et. al. J Neurotrauma. 2019 Dec 1;36(23):3233-3243.
Visual cognition in non-amnestic Alzheimer’s disease: Relations to tau, amyloid, and cortical atrophy
Heterogeneity within the Alzheimer’s disease (AD) syndromic spectrum is typically classified in a domain-specific manner (e.g., language vs. visual cognitive function). The central aim of this study was to investigate whether impairment in visual cognitive tasks thought to be subserved by posterior cortical dysfunction in non-amnestic AD presentations is associated with tau, amyloid, or neurodegeneration in those regions using 18F-AV-1451 and 11C-PiB positron emission tomography (PET) and magnetic resonance imaging (MRI).
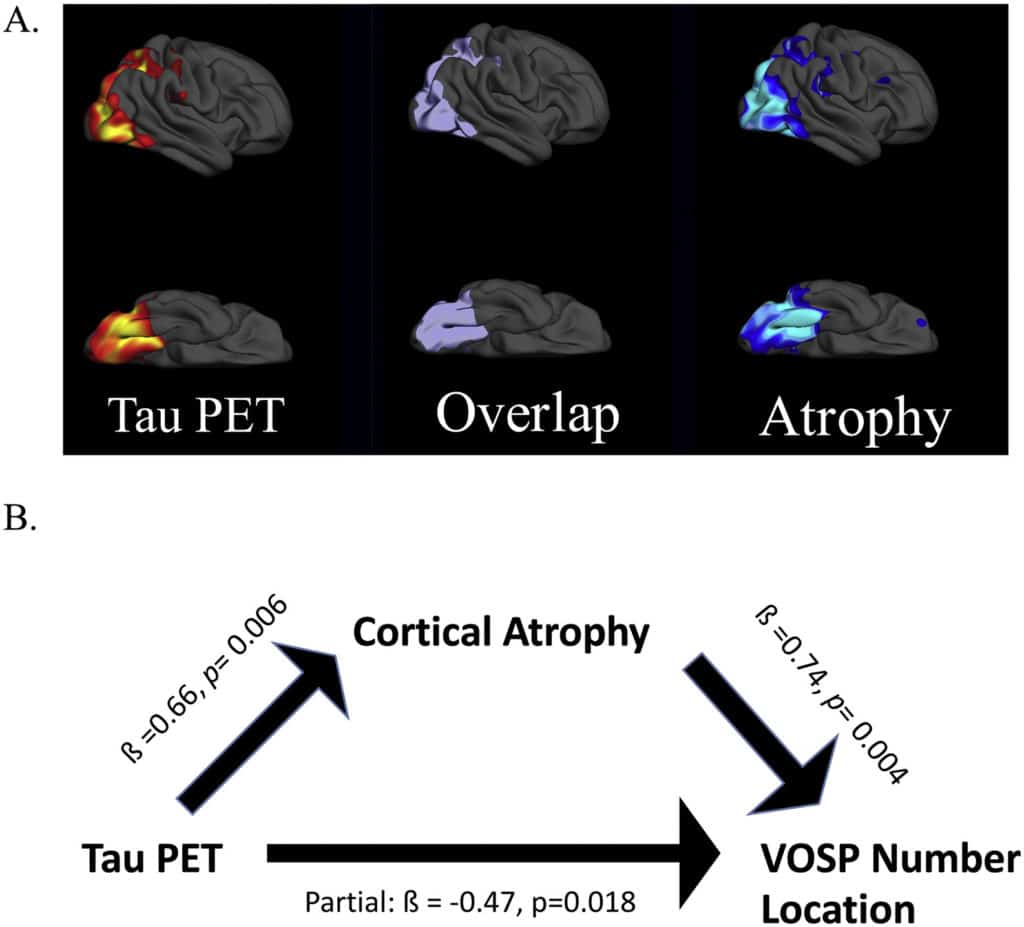
Sixteen amyloid-positive patients who met criteria for either Posterior Cortical Atrophy (PCA; n = 10) or logopenic variant Primary Progressive Aphasia (lvPPA; n = 6) were studied. All participants underwent a structured clinical assessment, neuropsychological battery, structural MRI, amyloid PET, and tau PET. The neuropsychological battery included two visual cognitive tests: VOSP Number Location and Benton Facial Recognition. Surface-based whole-cortical general linear models were used to first explore the similarities and differences between these biomarkers in the two patient groups, and then to assess their regional associations with visual cognitive test performance. The results show that these two variants of AD have both dissociable and overlapping areas of tau and atrophy, but amyloid is distributed with a stereotyped localization in both variants.
Performance on both visual cognitive tests were associated with tau and atrophy in the right lateral and medial occipital association cortex, superior parietal cortex, and posterior ventral occipitotemporal cortex. No cortical associations were observed with amyloid PET. We further demonstrate that cortical atrophy has a partially mediating effect on the association between tau pathology and visual cognitive task performance. Our findings show that non-amnestic variants of AD have partially dissociable spatial patterns of tau and atrophy that localize as expected based on symptoms, but similar patterns of amyloid. Further, we demonstrate that impairments of visual cognitive dysfunction are strongly associated with tau in visual cortical regions and mediated in part by atrophy.
D Putcha, et. al. Neuroimage Clin. 2019;23:101889. doi: 10.1016/j.nicl.2019.101889
Graph Convolutional Neural Networks for Alzheimer’s Disease Classification
Graph convolutional neural networks (GCNNs) aim to extend the data representation and classification capabilities of convolutional neural networks, which are highly effective for signals defined on regular Euclidean domains, e.g. image and audio signals, to irregular, graph-structured data defined on non-Euclidean domains. Graph-theoretic tools that enable us to study the brain as a complex system are of great significance in brain connectivity studies. Particularly, in the context of Alzheimer’s disease (AD), a neurodegenerative disorder associated with network dysfunction, graph-based tools are vital for disease classification and staging.
Here, we implement and test a multi-class GCNN classifier for network-based classification of subjects on the AD spectrum into four categories: cognitively normal, early mild cognitive impairment, late mild cognitive impairment, and AD. We train and validate the network using structural connectivity graphs obtained from diffusion tensor imaging data. Using receiver operating characteristic curves, we show that the GCNN classifier outperforms a support vector machine classifier by margins that are reliant on disease category. Our findings indicate that the performance gap between the two methods increases with disease progression from CN to AD. We thus demonstrate that GCNN is a competitive tool for staging and classification of subjects on the AD spectrum.
TA Song, et. al. Proc IEEE Int Symp Biomed Imaging. 2019 Apr;2019:414-417. doi: 10.1109/ISBI.2019.8759531. Epub 2019 Jul 11.
SANTIS: Sampling-Augmented Neural neTwork with Incoherent Structure for MR image reconstruction
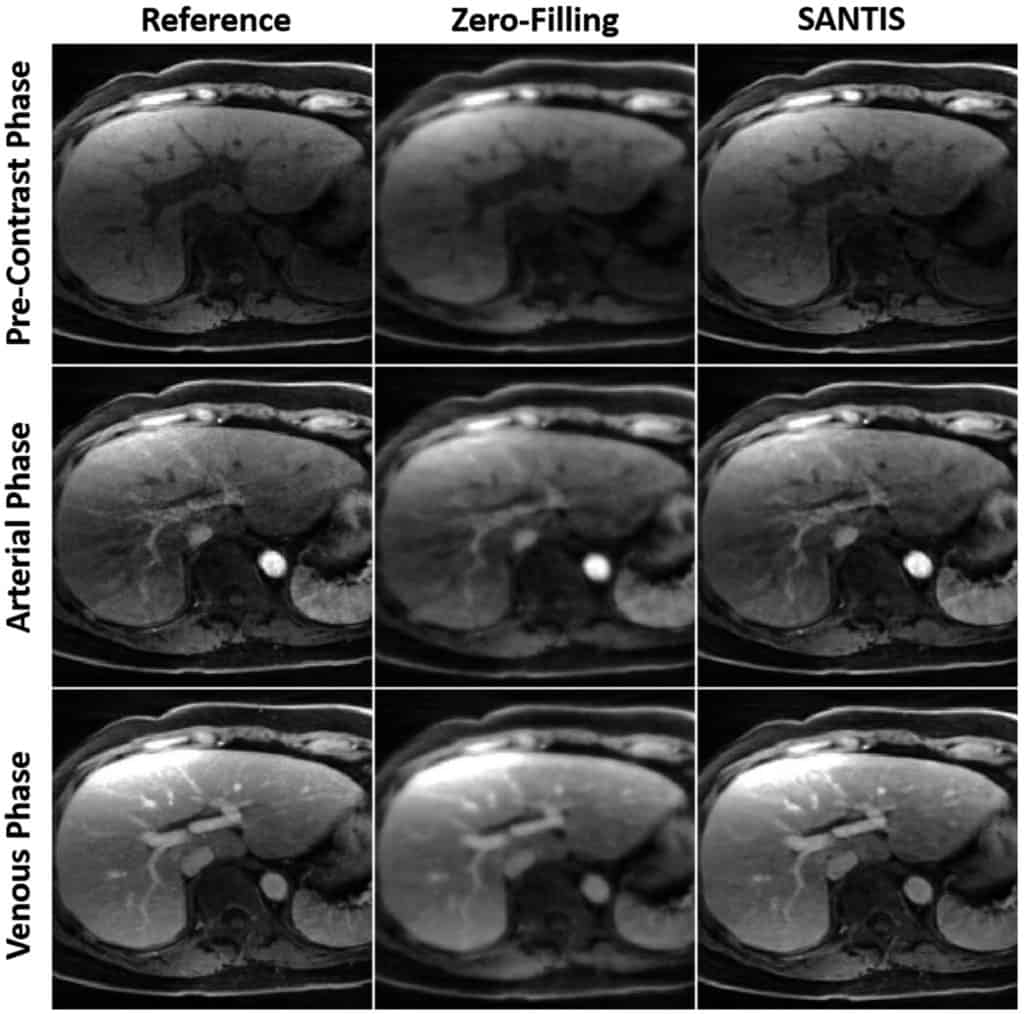
Purpose: To develop and evaluate a novel deep learning-based reconstruction framework called SANTIS (Sampling-Augmented Neural neTwork with Incoherent Structure) for efficient MR image reconstruction with improved robustness against sampling pattern discrepancy.
Methods: With a combination of data cycle-consistent adversarial network, end-to-end convolutional neural network mapping, and data fidelity enforcement for reconstructing undersampled MR data, SANTIS additionally utilizes a sampling-augmented training strategy by extensively varying undersampling patterns during training, so that the network is capable of learning various aliasing structures and thereby removing undersampling artifacts more effectively and robustly. The performance of SANTIS was demonstrated for accelerated knee imaging and liver imaging using a Cartesian trajectory and a golden-angle radial trajectory, respectively. Quantitative metrics were used to assess its performance against different references. The feasibility of SANTIS in reconstructing dynamic contrast-enhanced images was also demonstrated using transfer learning.
Results: Compared to conventional reconstruction that exploits image sparsity, SANTIS achieved consistently improved reconstruction performance (lower errors and greater image sharpness). Compared to standard learning-based methods without sampling augmentation (e.g., training with a fixed undersampling pattern), SANTIS provides comparable reconstruction performance, but significantly improved robustness, against sampling pattern discrepancy. SANTIS also achieved encouraging results for reconstructing liver images acquired at different contrast phases.
Conclusion: By extensively varying undersampling patterns, the sampling-augmented training strategy in SANTIS can remove undersampling artifacts more robustly. The novel concept behind SANTIS can particularly be useful for improving the robustness of deep learning-based image reconstruction against discrepancy between training and inference, an important, but currently less explored, topic.
F Liu, et. al. Magn Reson Med. 2019 Nov;82(5):1890-1904. doi: 10.1002/mrm.27827.
Computationally efficient deep neural network for computed tomography image reconstruction
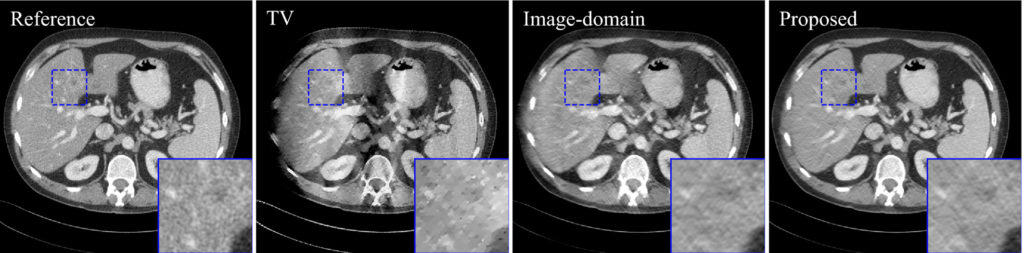
The reconstructed images of two‐dimensional 150 deg. limited‐angle study. Image‐domain result was the reconstructed images at unroll 1. TV result is provided to demonstrate the severity of limited‐angle artifacts. The display window is [−160,240] HU.
Purpose: Deep neural network-based image reconstruction has demonstrated promising performance in medical imaging for undersampled and low-dose scenarios. However, it requires large amount of memory and extensive time for the training. It is especially challenging to train the reconstruction networks for three-dimensional computed tomography (CT) because of the high resolution of CT images. The purpose of this work is to reduce the memory and time consumption of the training of the reconstruction networks for CT to make it practical for current hardware, while maintaining the quality of the reconstructed images.
Methods: We unrolled the proximal gradient descent algorithm for iterative image reconstruction to finite iterations and replaced the terms related to the penalty function with trainable convolutional neural networks (CNN). The network was trained greedily iteration by iteration in the image domain on patches, which requires reasonable amount of memory and time on mainstream graphics processing unit (GPU). To overcome the local-minimum problem caused by greedy learning, we used deep UNet as the CNN and incorporated separable quadratic surrogate with ordered subsets for data fidelity, so that the solution could escape from easy local minimums and achieve better image quality.
Results: The proposed method achieved comparable image quality with state-of-the-art neural network for CT image reconstruction on two-dimensional (2D) sparse-view and limited-angle problems on the low-dose CT challenge dataset. The difference in root-mean-square-error (RMSE) and structural similarity index (SSIM) was within [-0.23,0.47] HU and [0,0.001], respectively, with 95% confidence level. For three-dimensional (3D) image reconstruction with ordinary-size CT volume, the proposed method only needed 2 GB graphics processing unit (GPU) memory and 0.45 s per training iteration as minimum requirement, whereas existing methods may require 417 GB and 31 min. The proposed method achieved improved performance compared to total variation- and dictionary learning-based iterative reconstruction for both 2D and 3D problems.
Conclusions: We proposed a training-time computationally efficient neural network for CT image reconstruction. The proposed method achieved comparable image quality with state-of-the-art neural network for CT reconstruction, with significantly reduced memory and time requirement during training. The proposed method is applicable to 3D image reconstruction problems such as cone-beam CT and tomosynthesis on mainstream GPUs.
D Wu, et. al. Med Phys. 2019 Nov;46(11):4763-4776. doi: 10.1002/mp.13627.
Iterative PET Image Reconstruction Using Convolutional Neural Network Representation

PET image reconstruction is challenging due to the ill-poseness of the inverse problem and limited number of detected photons. Recently, the deep neural networks have been widely and successfully used in computer vision tasks and attracted growing interests in medical imaging. In this paper, we trained a deep residual convolutional neural network to improve PET image quality by using the existing inter-patient information. An innovative feature of the proposed method is that we embed the neural network in the iterative reconstruction framework for image representation, rather than using it as a post-processing tool. We formulate the objective function as a constrained optimization problem and solve it using the alternating direction method of multipliers algorithm. Both simulation data and hybrid real data are used to evaluate the proposed method. Quantification results show that our proposed iterative neural network method can outperform the neural network denoising and conventional penalized maximum likelihood methods.
K Gong, et. al. IEEE Trans Med Imaging. 2019 Mar;38(3):675-685. doi: 10.1109/TMI.2018.2869871.
Time of flight PET reconstruction using nonuniform update for regional recovery uniformity
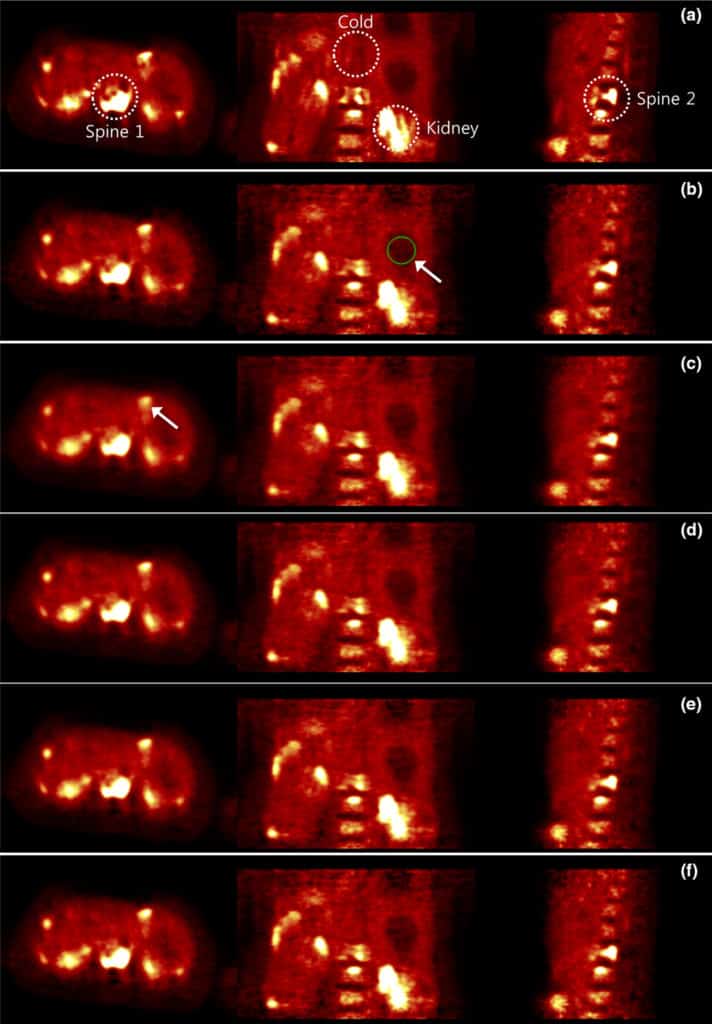
Purpose: Time of flight (TOF) PET reconstruction is well known to statistically improve the image quality compared to non-TOF PET. Although TOF PET can improve the overall signal to noise ratio (SNR) of the image compared to non-TOF PET, the SNR disparity between separate regions in the reconstructed image using TOF data becomes higher than that using non-TOF data. Using the conventional ordered subset expectation maximization (OS-EM) method, the SNR in the low activity regions becomes significantly lower than in the high activity regions due to the different photon statistics of TOF bins. A uniform recovery across different SNR regions is preferred if it can yield an overall good image quality within small number of iterations in practice. To allow more uniform recovery of regions, a spatially variant update is necessary for different SNR regions.
Methods: This paper focuses on designing a spatially variant step size and proposes a TOF-PET reconstruction method that uses a nonuniform separable quadratic surrogates (NUSQS) algorithm, providing a straightforward control of spatially variant step size. To control the noise, a spatially invariant quadratic regularization is incorporated, which by itself does not theoretically affect the recovery uniformity. The Nesterov’s momentum method with ordered subsets (OS) is also used to accelerate the reconstruction speed. To evaluate the proposed method, an XCAT simulation phantom and clinical data from a pancreas cancer patient with full (ground truth) and 6× downsampled counts were used, where a Poisson thinning process was employed for downsampling. We selected tumor and cold regions of interest (ROIs) and compared the proposed method with the TOF-based conventional OS-EM and OS-SQS algorithms with an early stopping criterion.
Results: In computer simulation, without regularization, hot regions of OS-EM and OS-NUSQS converged similarly, but cold region of OS-EM was noisier than OS-NUSQS after 24 iterations. With regularization, although the overall speeds of OS-EM and OS-NUSQS were similar, recovery ratios of hot and cold regions reconstructed by the OS-NUSQS were more uniform compared to those of the conventional OS-SQS and OS-EM. The OS-NUSQS with Nesterov’s momentum converged faster than others while preserving the uniform recovery. In the clinical example, we demonstrated that the OS-NUSQS with Nesterov’s momentum provides more uniform recovery ratios of hot and cold ROIs compared to the OS-SQS and OS-EM. Although the cost function of all methods is equivalent, the proposed method has higher structural similarity (SSIM) values of hot and cold regions compared to other methods after 24 iterations. Furthermore, our computing time using graphics processing unit was 80× shorter than the time using quad-core CPUs.
Conclusion: This paper proposes a TOF PET reconstruction method using the OS-NUSQS with Nesterov’s momentum for uniform recovery of different SNR regions. In particular, the spatially nonuniform step size in the proposed method provides uniform recovery ratios of different SNR regions, and the Nesterov’s momentum further accelerates overall convergence while preserving uniform recovery. The computer simulation and clinical example demonstrate that the proposed method converges uniformly across ROIs. In addition, tumor contrast and SSIM of the proposed method were higher than those of the conventional OS-EM and OS-SQS in early iterations.
K Kim, et. al. Med Phys. 2019 Feb;46(2):649-664. doi: 10.1002/mp.13321.
Penalized PET Reconstruction Using Deep Learning Prior and Local Linear Fitting
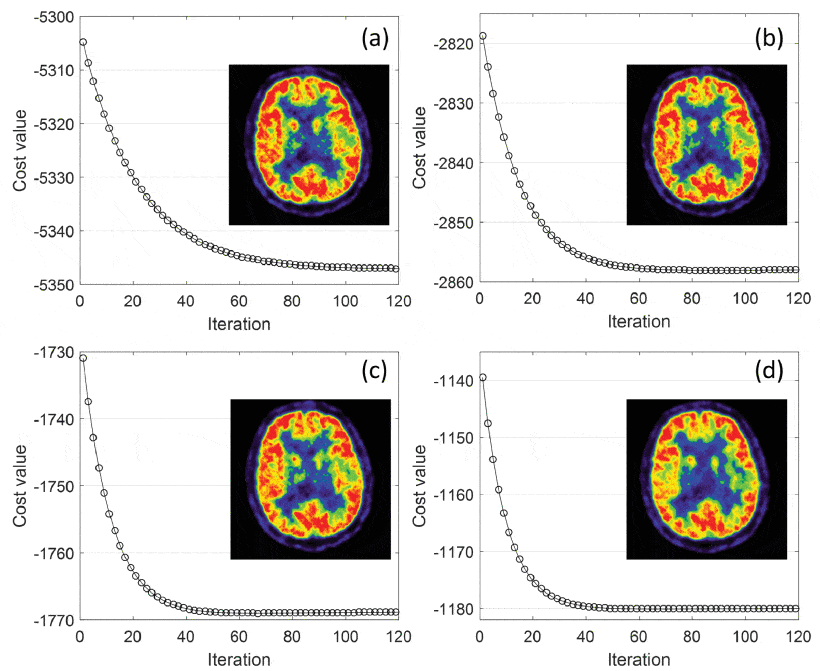
Motivated by the great potential of deep learning in medical imaging, we propose an iterative positron emission tomography reconstruction framework using a deep learning-based prior. We utilized the denoising convolutional neural network (DnCNN) method and trained the network using full-dose images as the ground truth and low dose images reconstructed from downsampled data by Poisson thinning as input. Since most published deep networks are trained at a predetermined noise level, the noise level disparity of training and testing data is a major problem for their applicability as a generalized prior. In particular, the noise level significantly changes in each iteration, which can potentially degrade the overall performance of iterative reconstruction.
Due to insufficient existing studies, we conducted simulations and evaluated the degradation of performance at various noise conditions. Our findings indicated that DnCNN produces additional bias induced by the disparity of noise levels. To address this issue, we propose a local linear fitting function incorporated with the DnCNN prior to improve the image quality by preventing unwanted bias. We demonstrate that the resultant method is robust against noise level disparities despite the network being trained at a predetermined noise level.
By means of bias and standard deviation studies via both simulations and clinical experiments, we show that the proposed method outperforms conventional methods based on total variation and non-local means penalties. We thereby confirm that the proposed method improves the reconstruction result both quantitatively and qualitatively.
K Kim, et. al. IEEE Trans Med Imaging. 2018 Jun;37(6):1478-1487. doi: 10.1109/TMI.2018.2832613.
A novel depth-of-interaction rebinning strategy for ultrahigh resolution PET
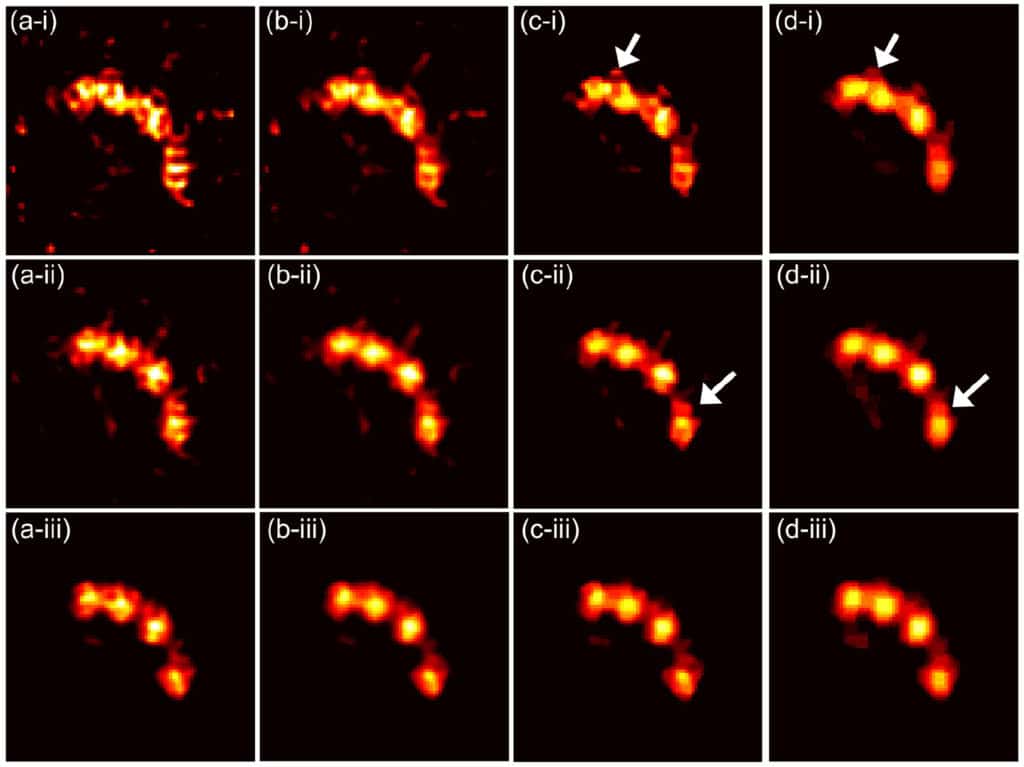
Small animal positron emission tomography (PET) imaging often requires high resolution (∼few hundred microns) to enable accurate quantitation in small structures such as animal brains. Recently, we have developed a prototype ultrahigh resolution depth-of-interaction (DOI) PET system that uses CdZnTe detectors with a detector pixel size of 350 μm and eight DOI layers with a 250 μm depth resolution. Due to the large number of line-of-response (LOR) combinations of DOIs, the system matrix for reconstruction is 64 times larger than that without DOI. While a high resolution virtual ring geometry can be employed to simplify the system matrix and create a sinogram, the LORs in such a sinogram tend to be sparse and irregular, leading to potential degradation of the reconstructed image quality.
In this paper, we propose a novel high resolution sinogram rebinning method in which a uniform sub-sampling DOI strategy is employed. However, even with the high resolution rebinning strategy, the reconstructed image tends to be very noisy due to insufficient photon counts in many high resolution sinogram pixels. To reduce noise effects, we developed a penalized maximum likelihood reconstruction framework with the Poisson log-likelihood and a non-convex total variation penalty. Here, an ordered subsets separable quadratic surrogate and alternating direction method of multipliers are utilized to solve the optimization. To evaluate the performance of the proposed sub-sampling method and the penalized maximum likelihood reconstruction technique, we perform simulations and preliminary point source experiments.
By comparing the reconstructed images and profiles based on sinograms without DOI, with rebinned DOI and with sub-sampled DOI, we demonstrate that the proposed method with sub-sampled DOIs can significantly improve the image quality with lower dose and yield a high resolution of <300 μm.
K Kim, et. al. Phys Med Biol. 2018 Aug 14;63(16):165011. doi: 10.1088/1361-6560/aad58c.
Low-dose CT reconstruction using spatially encoded nonlocal penalty
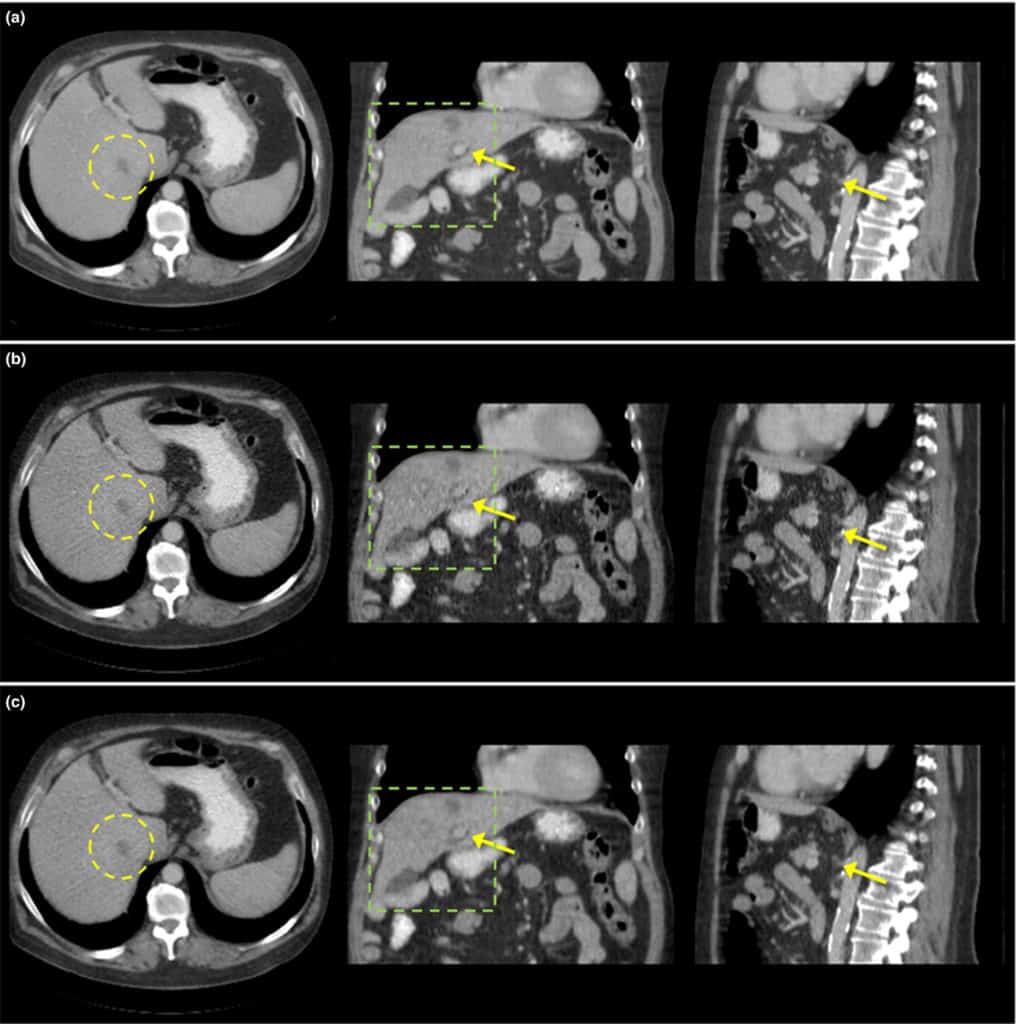
Purpose: Computed tomography (CT) is one of the most used imaging modalities for imaging both symptomatic and asymptomatic patients. However, because of the high demand for lower radiation dose during CT scans, the reconstructed image can suffer from noise and artifacts due to the trade-off between the image quality and the radiation dose. The purpose of this paper is to improve the image quality of quarter dose images and to select the best hyperparameters using the regular dose image as ground truth.
Methods: We first generated the axially stacked two-dimensional sinograms from the multislice raw projections with flying focal spots using a single slice rebinning method, which is an axially approximate method to provide simple implementation and efficient memory usage. To improve the image quality, a cost function containing the Poisson log-likelihood and spatially encoded nonlocal penalty is proposed. Specifically, an ordered subsets separable quadratic surrogates (OS-SQS) method for the log-likelihood is exploited and the patch-based similarity constraint with a spatially variant factor is developed to reduce the noise significantly while preserving features. Furthermore, we applied the Nesterov’s momentum method for acceleration and the diminishing number of subsets strategy for noise consistency. Fast nonlocal weight calculation is also utilized to reduce the computational cost.
Results: Datasets given by the Low Dose CT Grand Challenge were used for the validation, exploiting the training datasets with the regular and quarter dose data. The most important step in this paper was to fine-tune the hyperparameters to provide the best image for diagnosis. Using the regular dose filtered back-projection (FBP) image as ground truth, we could carefully select the hyperparameters by conducting a bias and standard deviation study, and we obtained the best images in a fixed number of iterations. We demonstrated that the proposed method with well selected hyperparameters improved the image quality using quarter dose data. The quarter dose proposed method was compared with the regular dose FBP, quarter dose FBP, and quarter dose l1 -based 3-D TV method. We confirmed that the quarter dose proposed image was comparable to the regular dose FBP image and was better than images using other quarter dose methods. The reconstructed test images of the accreditation (ACR) CT phantom and 20 patients data were evaluated by radiologists at the Mayo clinic, and this method was awarded first place in the Low Dose CT Grand Challenge.
Conclusion: We proposed the iterative CT reconstruction method using a spatially encoded nonlocal penalty and ordered subsets separable quadratic surrogates with the Nesterov’s momentum and diminishing number of subsets. The results demonstrated that the proposed method with fine-tuned hyperparameters can significantly improve the image quality and provide accurate diagnostic features at quarter dose. The performance of the proposed method should be further improved for small lesions, and a more thorough evaluation using additional clinical data is required in the future.
K Kim, et. al. Med Phys. 2017 Oct;44(10):e376-e390. doi: 10.1002/mp.12523.
Partial volume correction for PET quantification and its impact on brain network in Alzheimer’s disease
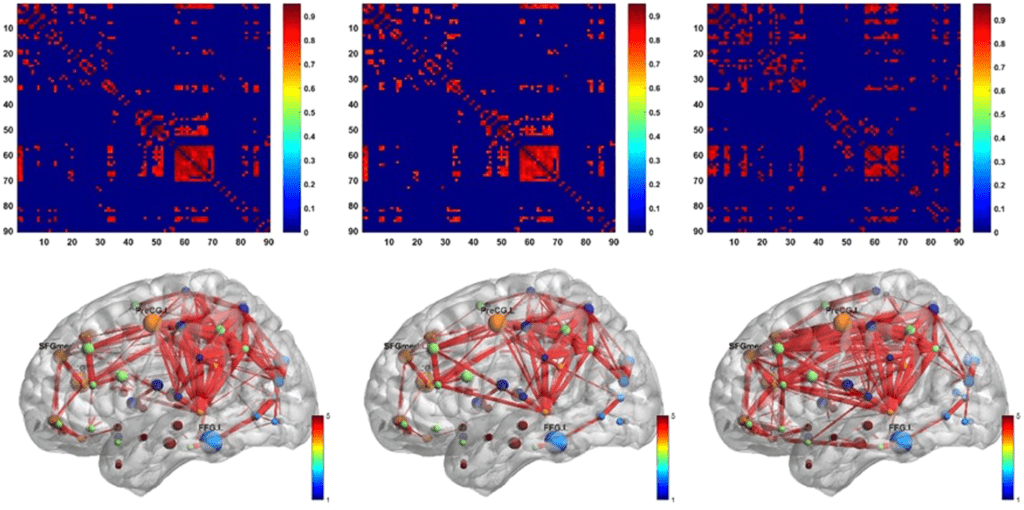
Amyloid positron emission tomography (PET) imaging is a valuable tool for research and diagnosis in Alzheimer’s disease (AD). Partial volume effects caused by the limited spatial resolution of PET scanners degrades the quantitative accuracy of PET image. In this study, we have applied a method to evaluate the impact of a joint-entropy based partial volume correction (PVC) technique on brain networks learned from a clinical dataset of AV-45 PET image and compare network properties of both uncorrected and corrected image-based brain networks. We also analyzed the region-wise SUVRs of both uncorrected and corrected images. We further performed classification tests on different groups using the same set of algorithms with same parameter settings. PVC has sometimes been avoided due to increased noise sensitivity in image registration and segmentation, however, our results indicate that appropriate PVC may enhance the brain network structure analysis for AD progression and improve classification performance.
J Yang, et. al. Sci Rep. 2017 Oct 12;7(1):13035. doi: 10.1038/s41598-017-13339-7.
Iterative Low-Dose CT Reconstruction With Priors Trained by Artificial Neural Network
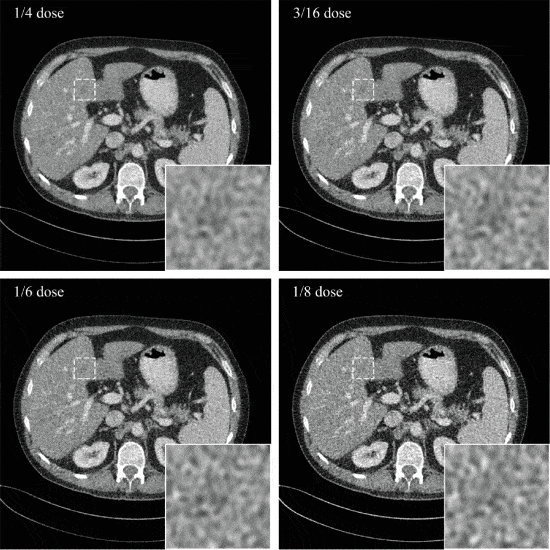
Dose reduction in computed tomography (CT) is essential for decreasing radiation risk in clinical applications. Iterative reconstruction algorithms are one of the most promising way to compensate for the increased noise due to reduction of photon flux. Most iterative reconstruction algorithms incorporate manually designed prior functions of the reconstructed image to suppress noises while maintaining structures of the image. These priors basically rely on smoothness constraints and cannot exploit more complex features of the image. The recent development of artificial neural networks and machine learning enabled learning of more complex features of image, which has the potential to improve reconstruction quality. In this letter, K-sparse auto encoder was used for unsupervised feature learning. A manifold was learned from normal-dose images and the distance between the reconstructed image and the manifold was minimized along with data fidelity during reconstruction. Experiments on 2016 Low-dose CT Grand Challenge were used for the method verification, and results demonstrated the noise reduction and detail preservation abilities of the proposed method.
D Wu, et. al. IEEE Trans Med Imaging. 2017 Dec;36(12):2479-2486. doi: 10.1109/TMI.2017.2753138.

